Barium »
PDB 1djh-2x6b »
2dns »
Barium in PDB 2dns: The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
Protein crystallography data
The structure of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine, PDB code: 2dns
was solved by
S.Okazaki,
A.Suzuki,
H.Komeda,
Y.Asano,
T.Yamane,
with X-Ray Crystallography technique. A brief refinement statistics is given in the table below:
| Resolution Low / High (Å) | 19.87 / 2.40 |
| Space group | P 1 21 1 |
| Cell size a, b, c (Å), α, β, γ (°) | 77.067, 123.702, 116.046, 90.00, 104.44, 90.00 |
| R / Rfree (%) | 19.5 / 26.9 |
Barium Binding Sites:
Pages:
>>> Page 1 <<< Page 2, Binding sites: 11 - 20;Binding sites:
The binding sites of Barium atom in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine (pdb code 2dns). This binding sites where shown within 5.0 Angstroms radius around Barium atom.In total 20 binding sites of Barium where determined in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine, PDB code: 2dns:
Jump to Barium binding site number: 1; 2; 3; 4; 5; 6; 7; 8; 9; 10;
Barium binding site 1 out of 20 in 2dns
Go back to
Barium binding site 1 out
of 20 in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
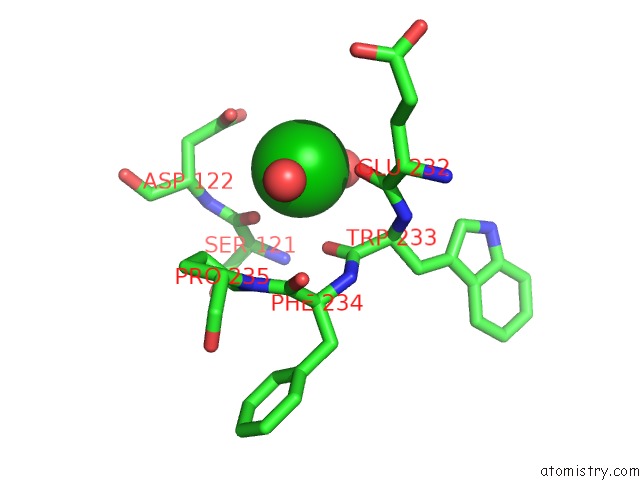
Mono view
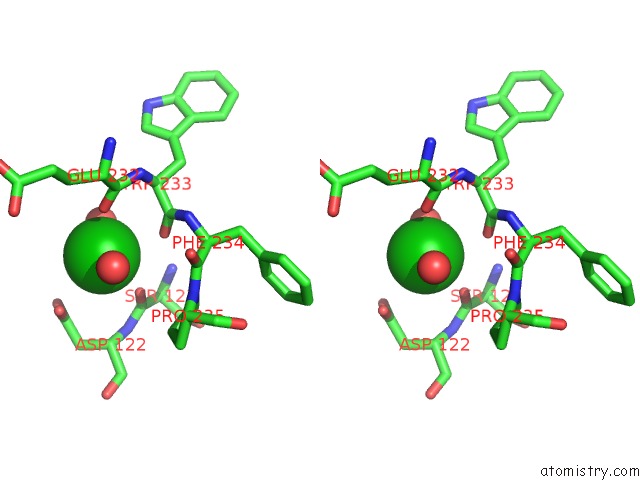
Stereo pair view

Mono view

Stereo pair view
A full contact list of Barium with other atoms in the Ba binding
site number 1 of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine within 5.0Å range:
|
Barium binding site 2 out of 20 in 2dns
Go back to
Barium binding site 2 out
of 20 in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
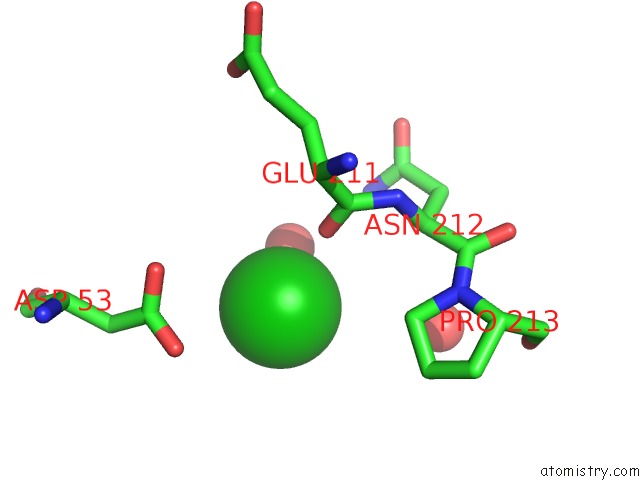
Mono view
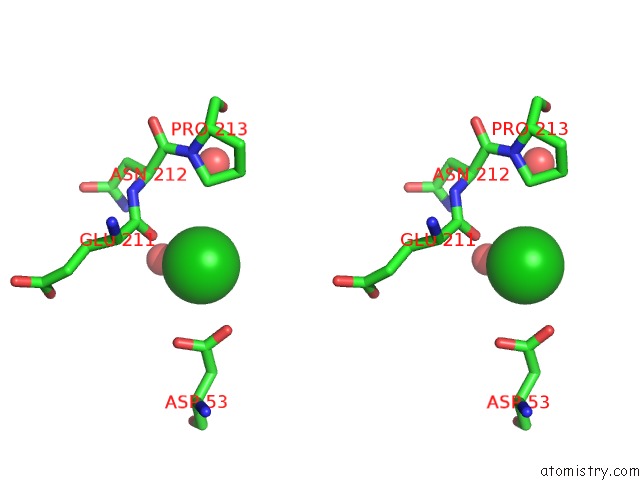
Stereo pair view

Mono view

Stereo pair view
A full contact list of Barium with other atoms in the Ba binding
site number 2 of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine within 5.0Å range:
|
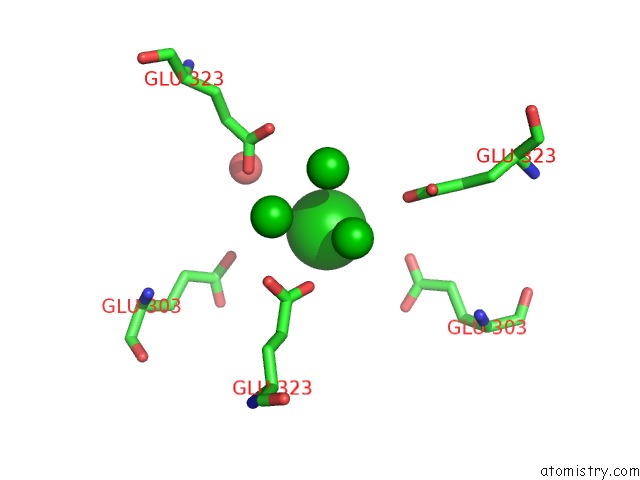
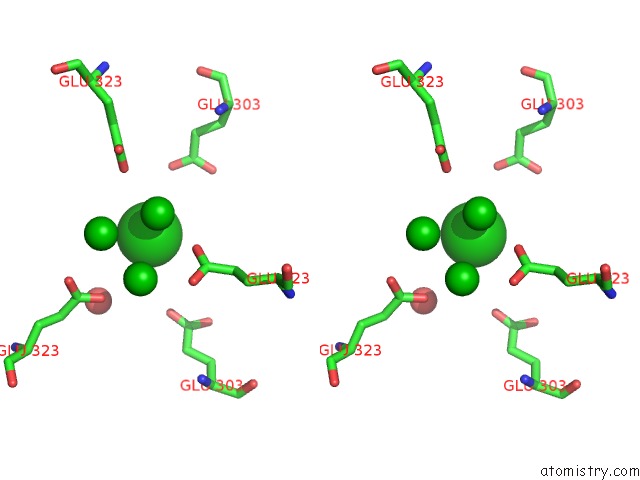
Barium binding site 3 out of 20 in 2dns
Go back to
Barium binding site 3 out
of 20 in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Barium with other atoms in the Ba binding
site number 3 of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine within 5.0Å range:
|
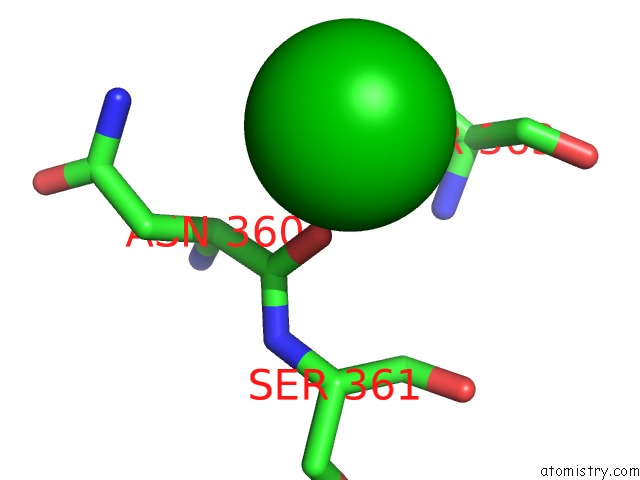
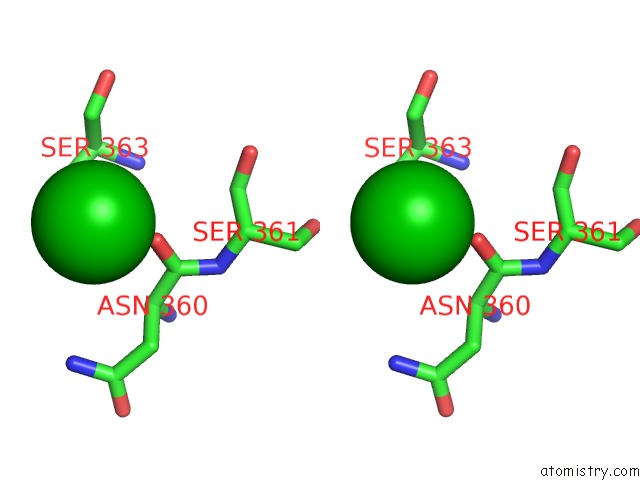
Barium binding site 4 out of 20 in 2dns
Go back to
Barium binding site 4 out
of 20 in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Barium with other atoms in the Ba binding
site number 4 of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine within 5.0Å range:
|
Barium binding site 5 out of 20 in 2dns
Go back to
Barium binding site 5 out
of 20 in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
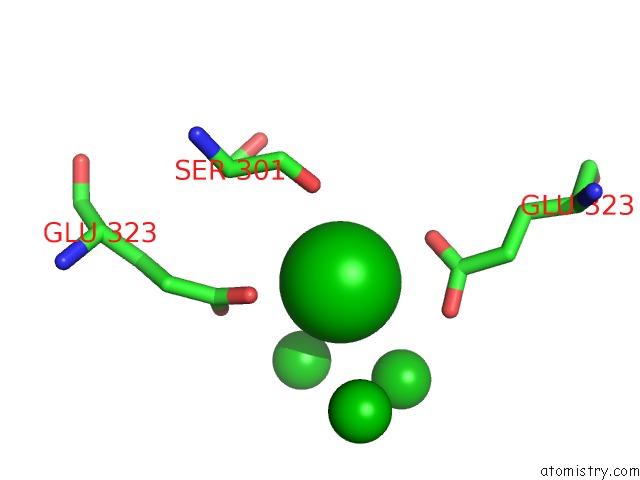
Mono view
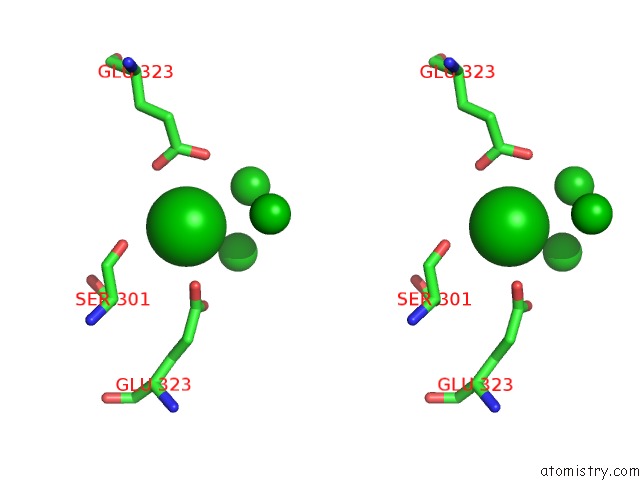
Stereo pair view

Mono view

Stereo pair view
A full contact list of Barium with other atoms in the Ba binding
site number 5 of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine within 5.0Å range:
|
Barium binding site 6 out of 20 in 2dns
Go back to
Barium binding site 6 out
of 20 in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
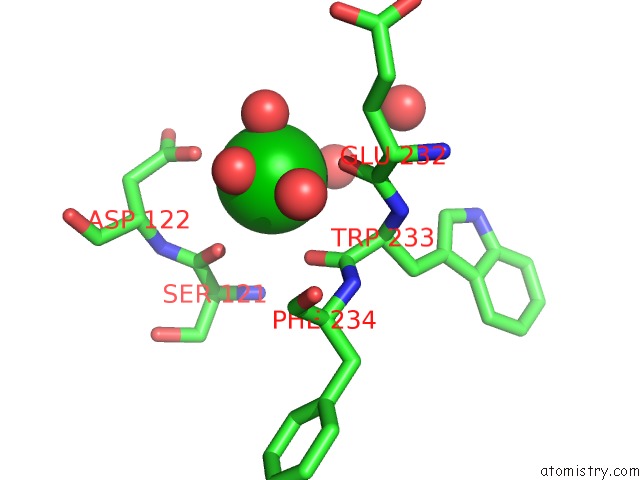
Mono view
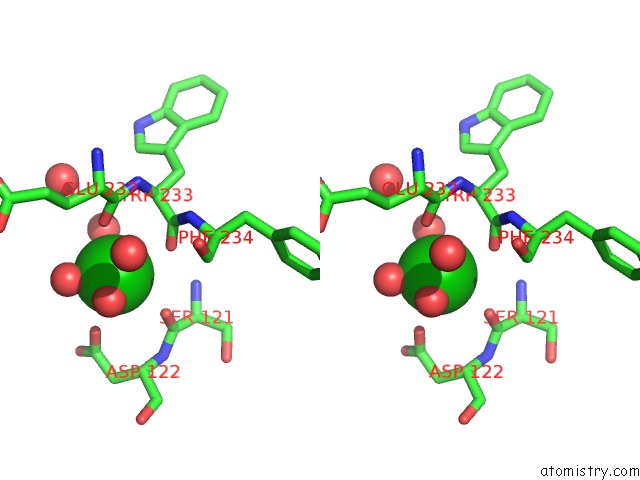
Stereo pair view

Mono view

Stereo pair view
A full contact list of Barium with other atoms in the Ba binding
site number 6 of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine within 5.0Å range:
|
Barium binding site 7 out of 20 in 2dns
Go back to
Barium binding site 7 out
of 20 in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
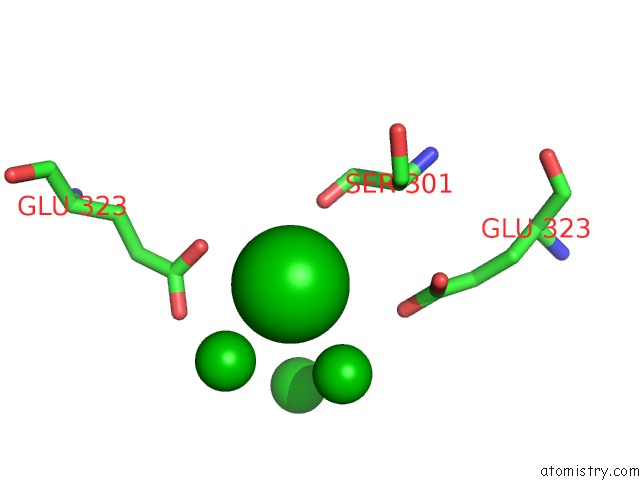
Mono view
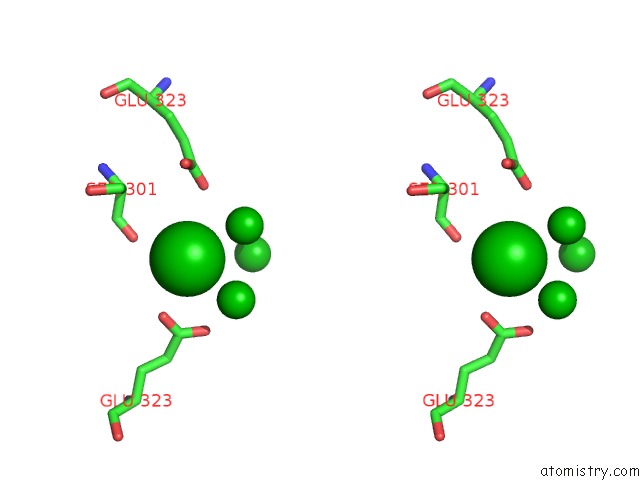
Stereo pair view

Mono view

Stereo pair view
A full contact list of Barium with other atoms in the Ba binding
site number 7 of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine within 5.0Å range:
|
Barium binding site 8 out of 20 in 2dns
Go back to
Barium binding site 8 out
of 20 in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
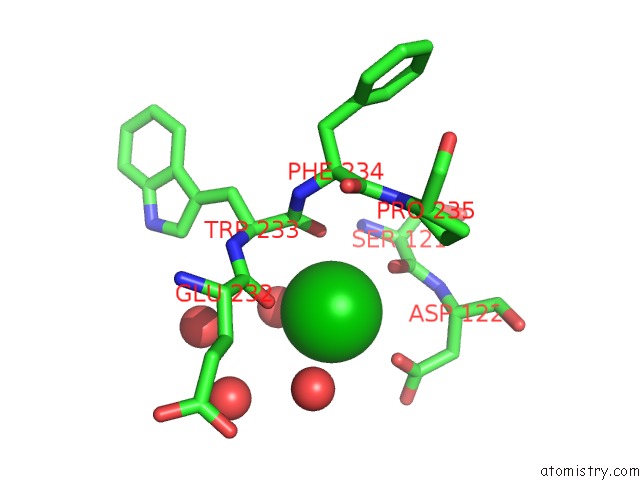
Mono view
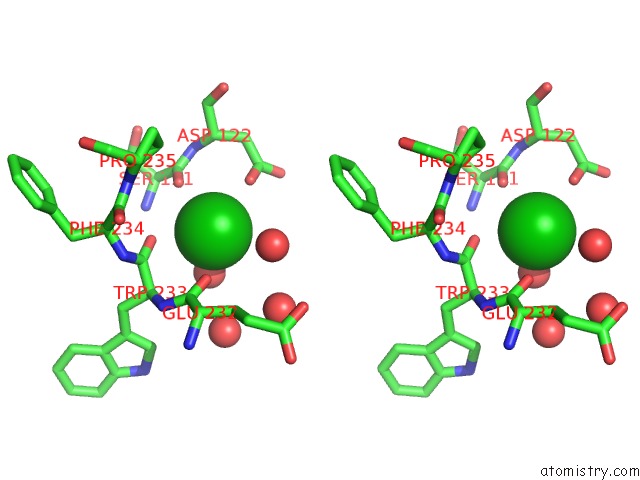
Stereo pair view

Mono view

Stereo pair view
A full contact list of Barium with other atoms in the Ba binding
site number 8 of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine within 5.0Å range:
|
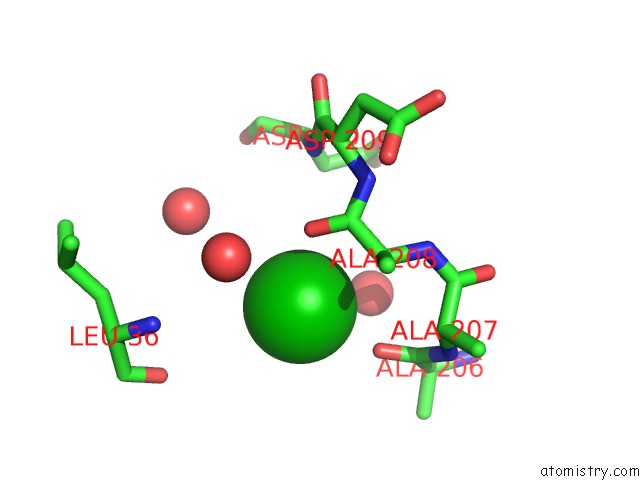
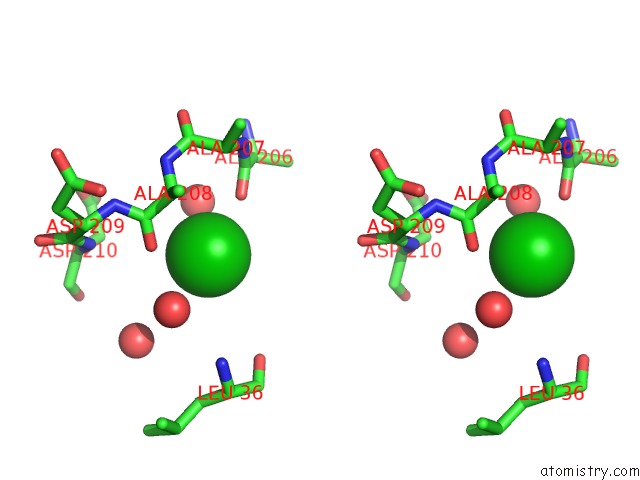
Barium binding site 9 out of 20 in 2dns
Go back to
Barium binding site 9 out
of 20 in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Barium with other atoms in the Ba binding
site number 9 of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine within 5.0Å range:
|
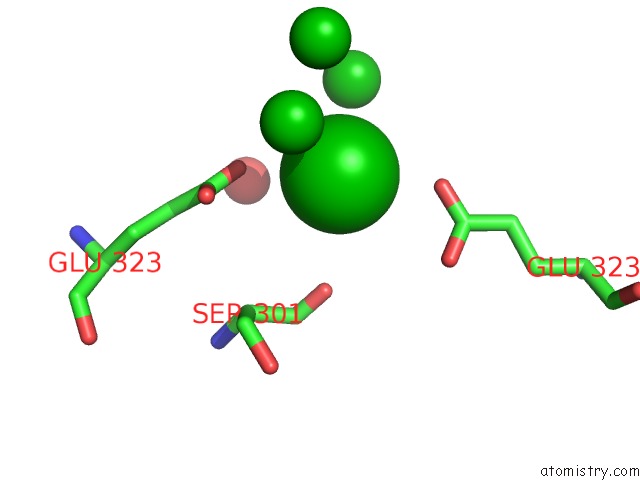
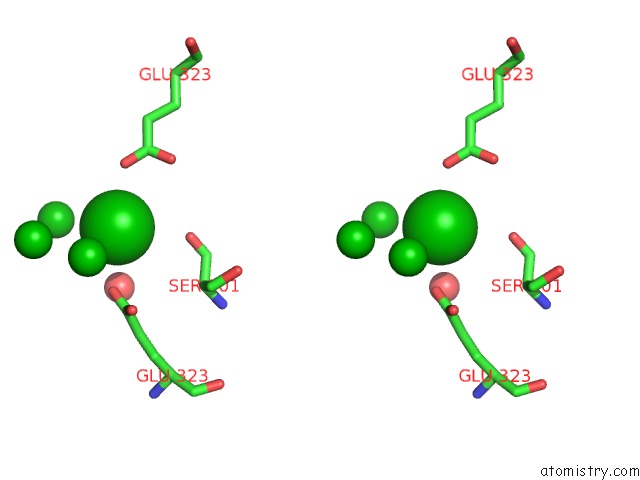
Barium binding site 10 out of 20 in 2dns
Go back to
Barium binding site 10 out
of 20 in the The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine
Mono view
Stereo pair view

Mono view

Stereo pair view
A full contact list of Barium with other atoms in the Ba binding
site number 10 of The Crystal Structure of D-Amino Acid Amidase From Ochrobactrum Anthropi SV3 Complexed with D-Phenylalanine within 5.0Å range:
|
Reference:
S.Okazaki,
A.Suzuki,
H.Komeda,
S.Yamaguchi,
Y.Asano,
T.Yamane.
Crystal Structure and Functional Characterization of A D-Stereospecific Amino Acid Amidase From Ochrobactrum Anthropi SV3, A New Member of the Penicillin-Recognizing Proteins J.Mol.Biol. V. 368 79 2007.
ISSN: ISSN 0022-2836
PubMed: 17331533
DOI: 10.1016/J.JMB.2006.10.070
Page generated: Wed Jul 10 15:01:51 2024
ISSN: ISSN 0022-2836
PubMed: 17331533
DOI: 10.1016/J.JMB.2006.10.070
Last articles
F in 4IVWF in 4IVY
F in 4IVO
F in 4IV4
F in 4IVM
F in 4IV2
F in 4IUI
F in 4IN4
F in 4IU7
F in 4ITI